
This process is called reverse transcription (RT), hence the name RT-PCR. To apply PCR to the study of RNA, the RNA sample must first be converted to cDNA to provide the necessary DNA template for the thermostable polymerase (Figure 1). Yet numerous instances exist in which amplification of RNA would be preferred. Thermostable DNA polymerases used for basic PCR require a DNA template, and as such, the technique is limited to the analysis of DNA samples. After 20–40 cycles, the amplified product may be analyzed for size, quantity, sequence, etc., or used in further experimental procedures. If the temperature during the annealing and extension steps are similar, these two steps can be combined into a single step in which both primer annealing and extension take place. The next cycle begins with a return to 94☌ for denaturation.Įach step of the cycle should be optimized for each template and primer pair combination. The extension step lasts approximately 1–2 minutes. For most thermostable DNA polymerases, this temperature is in the range of 70–74☌. Finally, the synthesis of new DNA begins as the reaction temperature is raised to the optimum for the DNA polymerase. This step lasts approximately 15–60 seconds. At this temperature, the oligonucleotide primers can form stable associations (anneal) with the denatured target DNA and serve as primers for the DNA polymerase. In the next step of a cycle, the temperature is reduced to approximately 40–60☌.
In the denaturation process, the two intertwined strands of DNA separate from one another, producing the necessary single-stranded DNA template for replication by the thermostable DNA polymerase. The initial step denatures the target DNA by heating it to 94☌ or higher for 15 seconds to 2 minutes. Ten cycles theoretically multiply the amplicon by a factor of about one thousand 20 cycles, by a factor of more than a million in a matter of hours.Įach cycle of PCR includes steps for template denaturation, primer annealing and primer extension. Each PCR cycle theoretically doubles the amount of targeted sequence (amplicon) in the reaction.
#TOUCHDOWN PCR TO AMPLIFY 3KB PLASMID REGION SERIES#
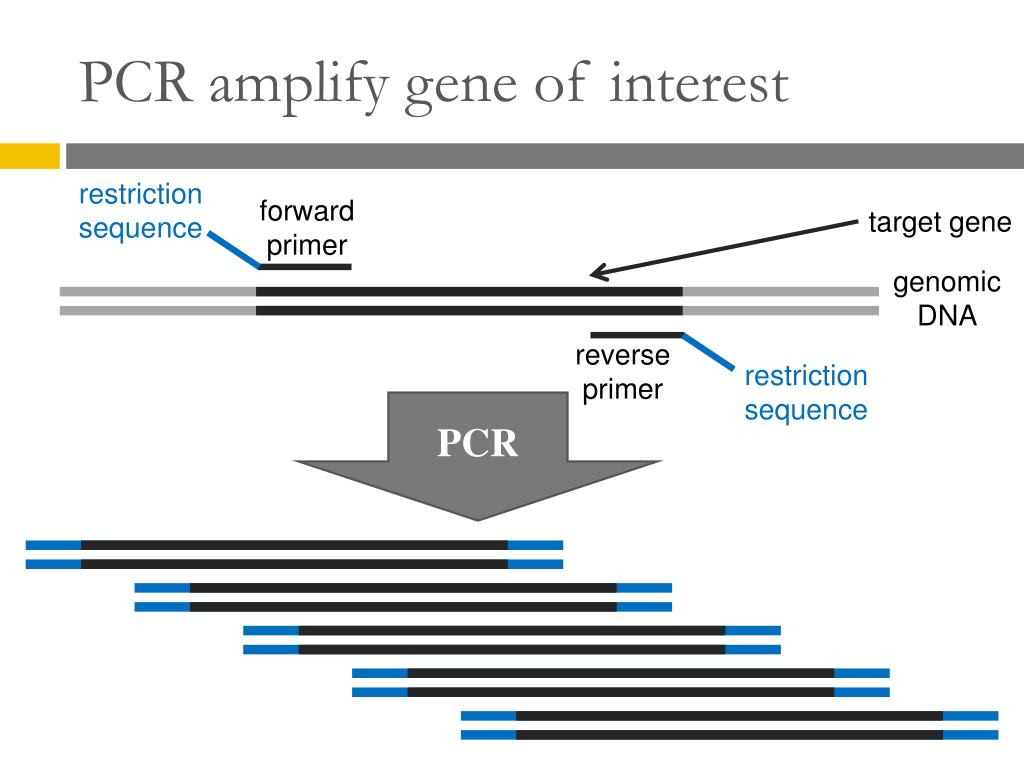
This series of temperature and time adjustments is referred to as one cycle of amplification. Once assembled, the reaction is placed in a thermal cycler, an instrument that subjects the reaction to a series of different temperatures for set amounts of time. A typical amplification reaction includes target DNA, a thermostable DNA polymerase, two oligonucleotide primers, deoxynucleotide triphosphates (dNTPs), reaction buffer and magnesium. The PCR process was originally developed to amplify short segments of a longer DNA molecule (Saiki et al. This chapter provides an overview of different types of PCR methods, applications and optimization. However, PCR has evolved far beyond simple amplification and detection, and many extensions of the original PCR method have been described. Basic PCR is commonplace in many molecular biology labs where it is used to amplify DNA fragments and detect DNA or RNA sequences within a cell or environment. These features make the technique extremely useful, not only in basic research, but also in commercial uses, including genetic identity testing, forensics, industrial quality control and in vitro diagnostics. Thus, PCR can achieve more sensitive detection and higher levels of amplification of specific sequences in less time than previously used methods. While most biochemical analyses, including nucleic acid detection with radioisotopes, require the input of significant amounts of biological material, the PCR process requires very little. Traditional methods of cloning a DNA sequence into a vector and replicating it in a living cell often require days or weeks of work, but amplification of DNA sequences by PCR requires only hours. The polymerase chain reaction (PCR) is a relatively simple technique that amplifies a DNA template to produce specific DNA fragments in vitro.


 0 kommentar(er)
0 kommentar(er)
